Design of Experiments¶
"The design of experiments (DOE) also known as experiment design or experimental design, is the design of any task that aims to describe and explain the variation of information under conditions that are hypothesized to reflect the variation" (see here). In other words, the DOE defines which experiments to perform in order to explore the parameter space of a given input/output problem.
In the frame of surrogate modelling for instance, the objective usually is to learn the behavior of an expensive black box function in order to cheaply predict a statistically coherent response to a given input. However, depending on the number of dimensions in the design space, exploration of the parameter space can quickly become overwhelming. Indeed, the uniform exploration of a parameter space grows exponentially with the number of dimensions which is often referred as the curse of dimensionality. For this reason, sophisticated sampling methods are often favoured over basic uniform sampling.
Design of Experiments in Melissa¶
In Melissa, the sampling relies on the parameter_generator server attribute which is set in the initializer of <use-case>_server.py. This object must be a function that yields an np.array() when called next(self.parameter_generator) containing the set of parameters for a single client (or group for Sobol Indices in the Sensitivity Analysis server). Melissa furnishes a set of parameter generators for users to choose from or modify, including a RandomUniform, HaltonSequence, LatinHypercube and Uniform3D. These all inherit from a base AbstractExperiment class which ensures the sampling is consistent. Additionally, the AbstractExperiment handles the creation of Sobol indices groups for the Sensitivity Analysis server if the user chooses to do so.
As shown in heat-pde (SA), the generator is instantiated as:
def __init__(self, config: Dict[str, Any]):
super().__init__(config)
self.nb_parms = self.study_options['nb_parameters']
Tmin, Tmax = self.study_options['parameter_range']
num_samples = self.study_options['num_samples']
# Example of random uniform sampling
self.parameter_generator = RandomUniform(
nb_parms=self.nb_parms, nb_sim=num_samples,
l_bounds=[Tmin], u_bounds=[Tmax],
second_order=False,
apply_pick_freeze=self.sobol_op
).generator()
Where the user is providing the foundation for the experimental design to the RandomUniform class including the number of parameters, the number of samples, and the bounds for each parameter to sample from. These methods can be tweaked for customization, as shown in heatpde_dl_server.py:
class RandomUniformHPDE(RandomUniform):
def __init__(self, mesh_size: int = 0, time_discretization: int = 0, **kwargs):
super().__init__(**kwargs)
self.mesh_size = mesh_size
self.time_discretization = time_discretization
def draw(self):
param_set = [self.mesh_size, self.mesh_size, self.time_discretization]
for n, _ in enumerate(range(self.nb_parms)):
param_set.append(random.uniform(self.l_bounds[n], self.u_bounds[n]))
# return param_set
return np.array(param_set)
Where we inherited from the base RandomUniform class to modify how the parameters are drawn and compiled for each client.
Uniform sampling¶
In the context of Melissa, users are mostly concerned with ensemble runs where, regardless of the number of dimensions, the number of computable solutions is often high enough for uniform sampling to stay relevant. Furthermore, When doing sensitivity analysis for instance, uniform sampling is particularly useful since it is an unbiased method that provides uncorrelated samples as required by the pick-freeze method.
Scipy based sampling¶
When dealing with deep surrogates, uniform sampling may however lead to slower learning due to a less efficient coverage of the design space. In addition, an unsatisfactory training may require the study to be pushed further. In this case, incremental parameter space search can be performed efficiently thanks to sequence sampling methods provided by the scipy.stats.qmc submodule.
Halton sequence¶
The Halton sequence is a deterministic sampling method. In Melissa, the HaltonGenerator is based on scipy.stats.qmc Halton sampler. The user can instantiate Melissa's built in HaltonGenerator with:
# Example of Halton sampling
self.parameter_generator = HaltonGenerator(nb_parms=self.nb_parms, nb_sim=num_samples,
l_bounds=[Tmin],u_bounds=[Tmax],second_order=False,
apply_pick_freeze=self.sobol_op).generator()
Latin Hypercube Sampling (LHS)¶
The latin hypercube sampling is a non-deterministic method. In Melissa, the LHSGenerator is based on scipy.stats.qmc Latin Hypercube sampler.
Note
Non-deterministic generators take a seed integer as argument in order to enforce the reproducibility of the generated inputs.
Warning
As opposed to the Halton sequence, sampling twice 10 samples from an LHS sampler won't yield the same DOE as when sampling 20 samples at once.
The user can instantiate the Melissa provided LHSGenerator with:
# Example of Latin Hypercube Sampling
self.parameter_generator = LHSGenerator(nb_parms=self.nb_parms, nb_sim=num_samples,
l_bounds=[Tmin],u_bounds=[Tmax],second_order=False,
apply_pick_freeze=self.sobol_op).generator()
DOE quality metrics¶
The Figure below compares the DOEs obtained with uniform, LHS and Halton sampling of 50 points across a parameter space of 2 dimensions:
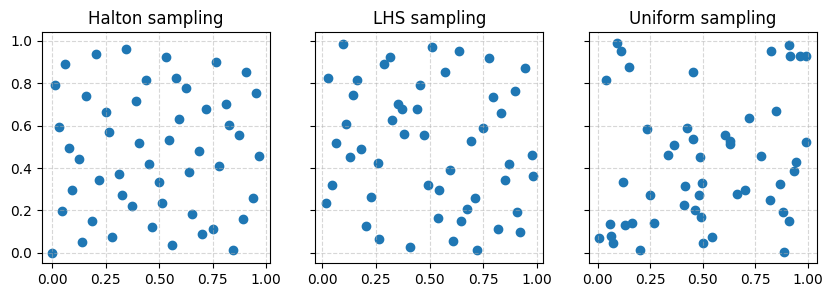
It clearly shows how uniform sampling may result in both cluttered and under-explored regions across the parameter space while Halton and LHS sampling provide a more homogeneous coverage.
In addition, as discussed earlier, LHS and Halton sampling are sequence samplers which means that their DOE can be enhanced a posteriori by resampling from the same generator. This feature is illustrated on the figure below where 20 points are added to the previous sets of parameters.
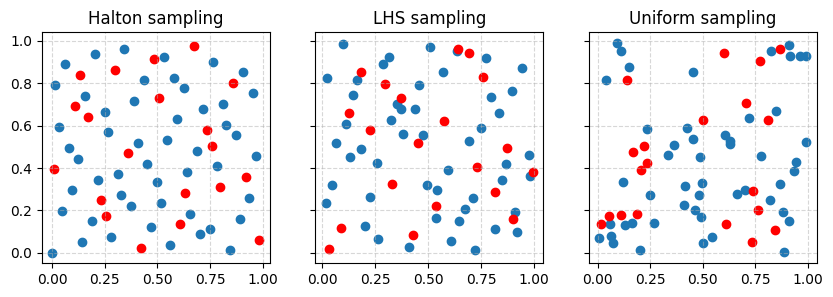
Finally, although the quality of the DOE may seem evident from the figures, intuition may be misleading. In order to evaluate the quality of a DOE, scipy.qmc comes with a discrepancy method:
The discrepancy is a uniformity criterion used to assess the space filling of a number of samples in a hypercube. A discrepancy quantifies the distance between the continuous uniform distribution on a hypercube and the discrete uniform distribution on distinct sample points.
The lower the value is, the better the coverage of the parameter space is.
For the DOEs represented in this section, the following discrepancies were obtained:
| Sampling | Sample size | Discrepancy |
|---|---|---|
| Uniform | 50 | 0.01167 |
| Uniform | 50+20 | 0.01045 |
| LHS | 50 | 0.00054 |
| LHS | 50+20 | 0.00041 |
| Halton | 50 | 0.00183 |
| Halton | 50+20 | 0.00097 |